Molecular Biology Techniques: Gene Therapy and cDNA Library Creation
Gene Therapy: Concepts and Applications
Ever since the dawn of mankind, diseases have plagued humans over the ages. Years of innovations and advancements in science have provided us with a deeper understanding of how diseases work. This has led to lower mortality rates and longer lifespans. But there are some diseases that just cannot be cured using traditional medicine or surgery. Gene therapy is an experimental technique that caters to patients with such diseases.
What is Gene Therapy?
Gene therapy is a technique which involves the replacement of defective genes with healthy ones in order to treat genetic disorders. It is an artificial method that introduces DNA into the cells of the human body. The first gene therapy was successfully accomplished in the year 1989.
The simple process of gene therapy is shown in the figure below:
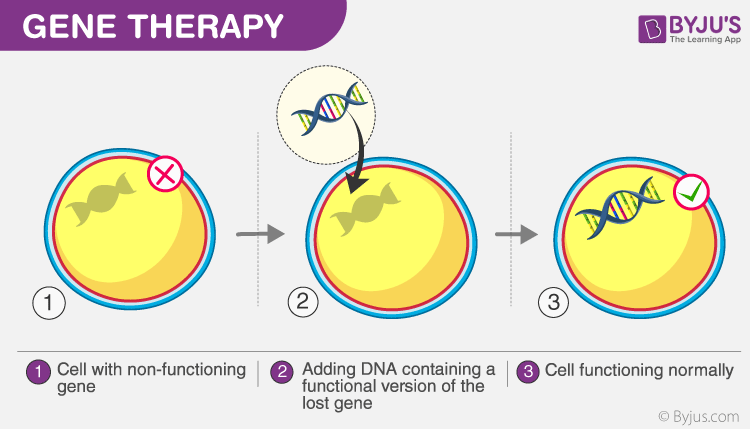
In the figure, the cell with the defective gene is injected with a normal gene, which helps in the normal functioning of the cell. This technique is employed mainly to fight against diseases in the human body and also to treat genetic disorders. The damaged proteins are replaced in the cell by the insertion of DNA into that cell. Generally, improper protein production in the cell leads to diseases. These diseases are treated using a gene therapy technique. For example, cancer cells contain faulty cells that are different from normal cells and have defective proteins. If these proteins are not replaced, this disease could prove fatal.
Types of Gene Therapy
Basically, there are two types of gene therapy:
Somatic Gene Therapy
This type usually occurs in the somatic cells of the human body. This is related to a single person, and only the person who has the damaged cells will receive the replacement with healthy cells. In this method, therapeutic genes are transferred into the somatic cells or the stem cells of the human body. This technique is considered the best and safest method of gene therapy.
Germline Gene Therapy
It occurs in the germline cells of the human body. Generally, this method is adopted to treat genetic, disease-causing variations of genes that are passed from parents to their children. The process involves introducing a healthy DNA into the cells responsible for producing reproductive cells (eggs or sperm). Germline gene therapy is not legal in many places as the risks outweigh the rewards.
Application of Gene Therapy
Gene therapy is utilized for several purposes:
- It is used in the replacement of genes that cause medical ill-health.
- The method generally destroys the problem-causing genes.
- It helps the body to fight against diseases by adding genes to the human body.
- This method is employed to treat diseases such as cancer, ADA deficiency, and cystic fibrosis, among others.
For more information about gene therapy, login to BYJU’S.
cDNA Library Construction and Methodology
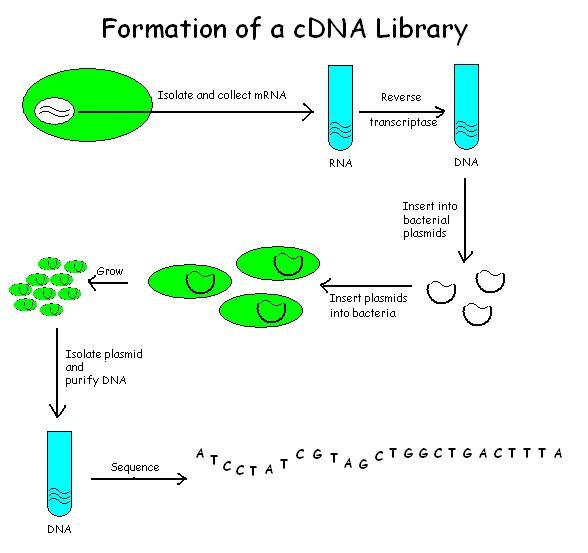
A cDNA library is a collection of cloned cDNA (complementary DNA) fragments inserted into host cells, representing a portion of the organism’s transcriptome, and stored as a “library.” cDNA is produced from fully transcribed mRNA found in the nucleus and therefore contains only the expressed genes of an organism. Similarly, tissue-specific cDNA libraries can be produced. In eukaryotic cells, the mature mRNA is already spliced; hence, the cDNA produced lacks introns and can be readily expressed in a bacterial cell.[1] While information in cDNA libraries is a powerful and useful tool since gene products are easily identified, the libraries lack information about enhancers, introns, and other regulatory elements found in a genomic DNA library.[2]
cDNA Library Construction Steps
cDNA is created from mature mRNA from a eukaryotic cell using reverse transcriptase. In eukaryotes, a poly-(A) tail (consisting of a long sequence of adenine nucleotides) distinguishes mRNA from tRNA and rRNA and can therefore be used as a primer site for reverse transcription. This presents a challenge because not all transcripts, such as those for histones, encode a poly-A tail.[3][4]
mRNA Extraction
Firstly, the mRNA template needs to be isolated for the creation of cDNA libraries. Since mRNA only contains exons, the integrity of the isolated mRNA should be considered to ensure the encoded protein can still be produced. Isolated mRNA should range from 500 bp to 8 kb.[5] Several methods exist for purifying RNA, such as trizol extraction and column purification. Column purification can be done using oligomeric dT nucleotide coated resins. Features of mRNA, such as having a poly-A tail, can be exploited, ensuring only mRNA sequences containing this feature will bind. The desired mRNA bound to the column is then eluted.
cDNA Construction
Once mRNA is purified, an oligo-dT primer (a short sequence of deoxy-thymidine nucleotides) is bound to the poly-A tail of the RNA. The primer is required to initiate DNA synthesis by the enzyme reverse transcriptase. This results in the creation of RNA-DNA hybrids, where a single strand of complementary DNA is bound to a strand of mRNA.[6] To remove the mRNA, the *RNase H* enzyme is used to cleave the backbone of the mRNA and generate free 3′-OH groups, which is important for the replacement of mRNA with DNA.[5] DNA polymerase I is then added; the cleaved RNA acts as a primer that DNA polymerase I can identify, initiating the replacement of RNA nucleotides with DNA nucleotides.[5] This is provided by the *ssDNA* itself by coiling on itself at the 3′ end, generating a hairpin loop. The polymerase extends the 3′-OH end, and later the loop at the 3′ end is opened by the scissoring action of S1 nuclease. Restriction endonucleases and DNA ligase are then used to clone the sequences into bacterial plasmids.
The cloned bacteria are then selected, commonly through the use of antibiotic selection. Once selected, stocks of the bacteria are created, which can later be grown and sequenced to compile the cDNA library.
cDNA Library Uses
cDNA libraries are commonly used when reproducing eukaryotic genomes, as the amount of information is reduced to remove the large numbers of non-coding regions from the library. cDNA libraries are used to express eukaryotic genes in prokaryotes. Prokaryotes do not have introns in their DNA and therefore do not possess the enzymes necessary to splice them out during the transcription process. cDNA does not have introns and therefore can be expressed in prokaryotic cells. cDNA libraries are most useful in *reverse genetics*, where the additional genomic information is of less use. Additionally, cDNA libraries are frequently used in *functional cloning* to identify genes based on the encoded protein’s function. When studying eukaryotic DNA, expression libraries are constructed using complementary DNA (cDNA) to help ensure the insert is truly a gene.[6]
Cloning of cDNA
cDNA molecules can be cloned by using restriction site linkers. Linkers are short, double-stranded pieces of DNA (*oligodeoxyribonucleotides*) about 8 to 12 nucleotide pairs long that include a restriction endonuclease cleavage site (e.g., *BamHI*). Both the cDNA and the linker have blunt ends, which can be ligated together using a high concentration of T4 DNA ligase. Sticky ends are then produced in the cDNA molecule by cleaving the cDNA ends (which now have linkers with an incorporated site) with the appropriate endonuclease. A cloning vector (plasmid) is then also cleaved with the appropriate endonuclease. Following “sticky end” ligation of the insert into the vector, the resulting recombinant DNA molecule is transferred into an *E. coli* host cell for cloning.
